Myogenesis & REGeneration Portal


Retrieving intracellular pathways
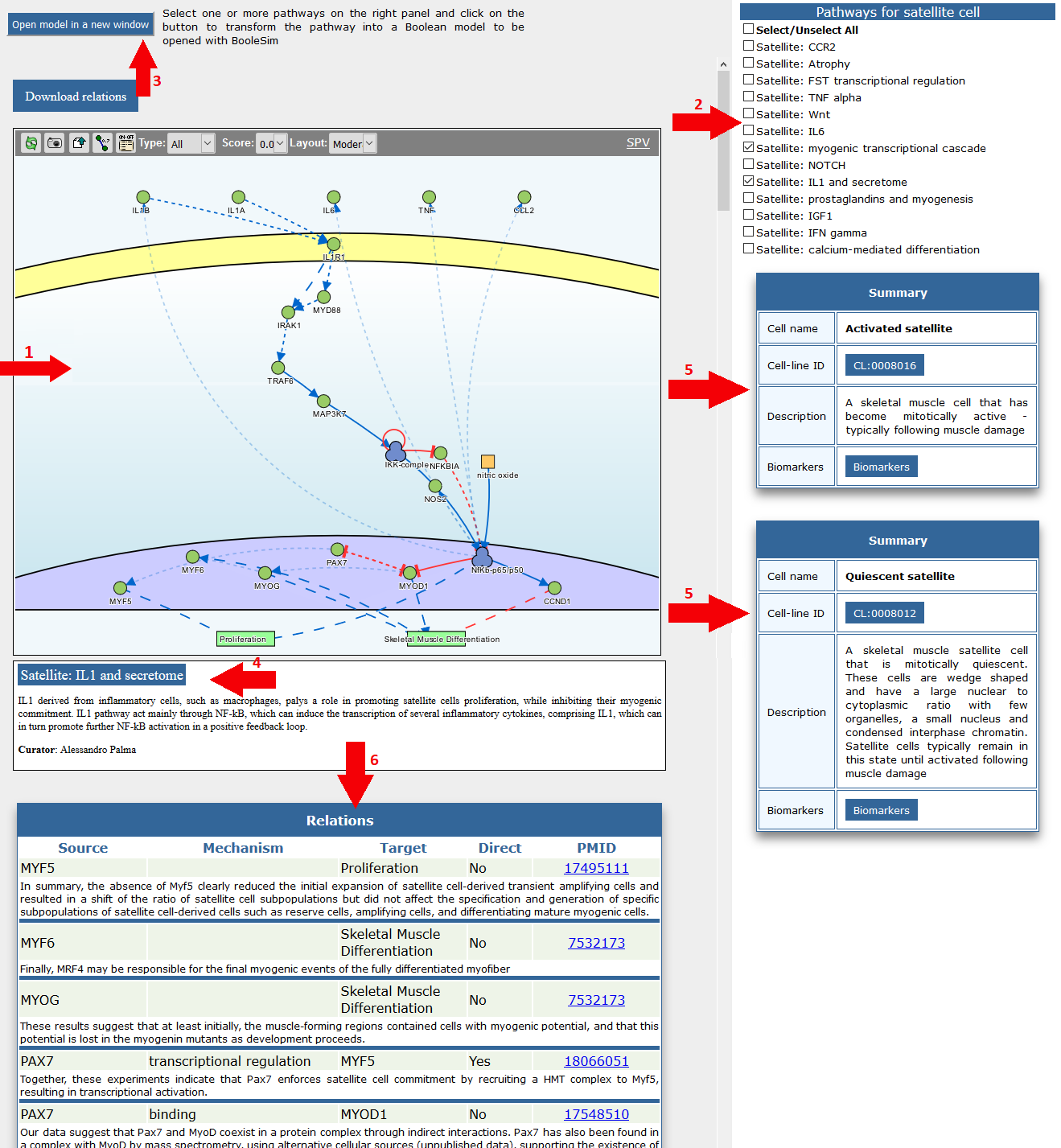
(1) Pathways are displayed in the cell type page via an interactive graph viewer where nodes and signed directed edges are clickable and linked to relevant informaton.
(2) Representative pathways for the selected cell type are listed on the right. User can select and merge two or more of these pathways by clicking check boxes.
(3) The “Build and open boolean model” button assembles a boolean model from any logic model displayed in the viewer and opens it in a Boolesim window where the model can be executed starting from different conditions.
For each pathway a short description (4) is provided below the interactive graph viewer to help the user to understand the relevance of that pathway for the differentiation decisions of the selected cell.
A summary description (5) of the selected cell is also displayed on the right pane.
Molecular interactions between signaling molecules are also reported as a table (6), to facilitate the retrieval of information on each reported interaction.
Exploring cell-cell interactions
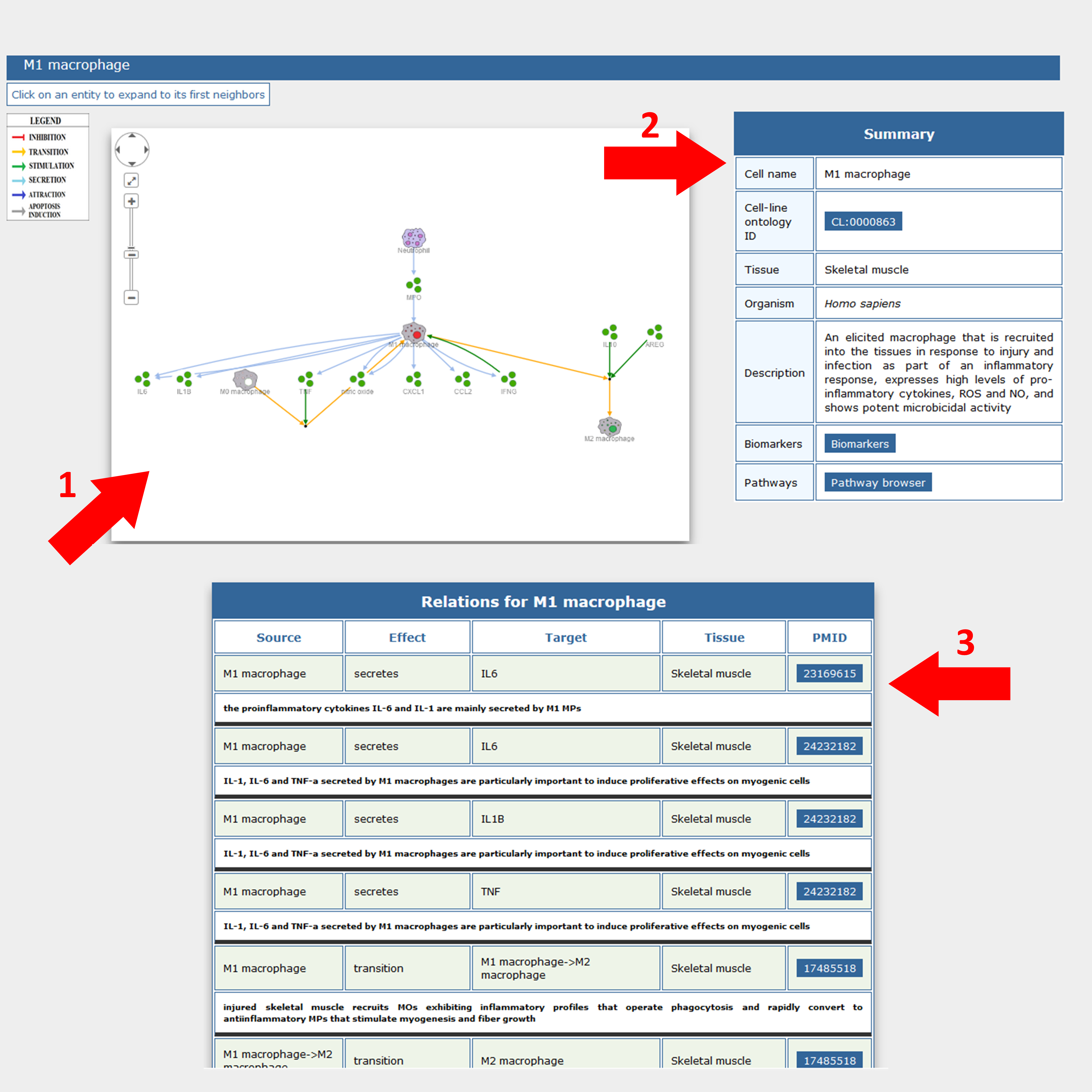
Cell-cell interactions are displayed via a cytoscape.js implementation (1) in the form of a signed directed graph where nodes and edges are clickable permitting the retrieval of information about entities (both cellular and molecular) and their relationships.
A short summary of the selected cell (2) on the right and a table of the cell relationships (3), at the bottom of the graph, are also provided.
The cell interaction graph can be expanded by clicking on graph nodes, either cell types or secreted signaling molecules that mediate cell interactions. This action expands the displayed network by adding the first neighbors of the selected entity. By a similar approach nodes can also be removed to simplify the graph representation.
Getting cell-specific biomarkers
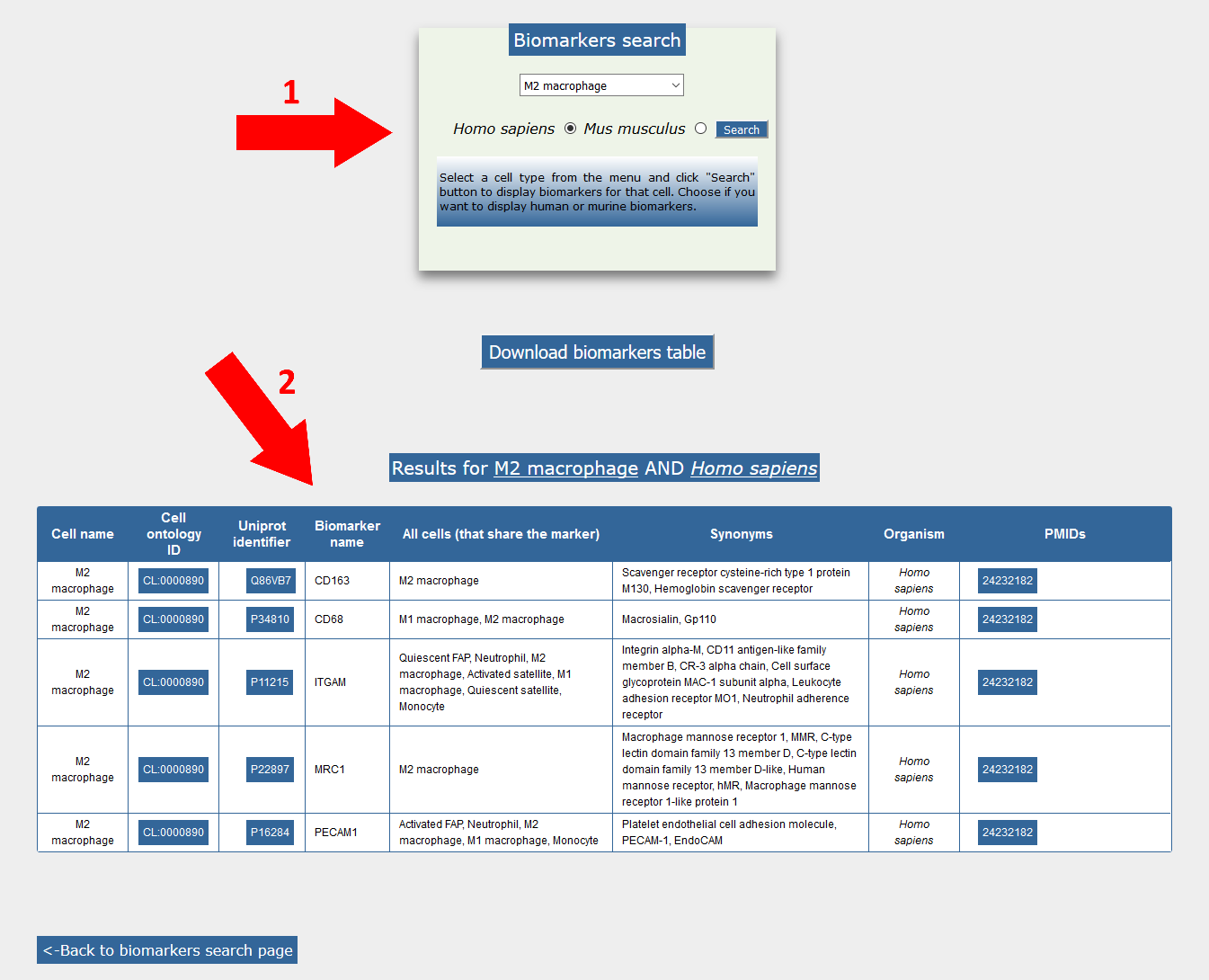
The biomarker page offers access to information about antigens that have been observed on the membrane of the different mononuclear cell type (1). A drop-down menu offers the possibility to select a cell type and directs to a table (2), containing information related to the selected cell biomarkers.
Downloading data
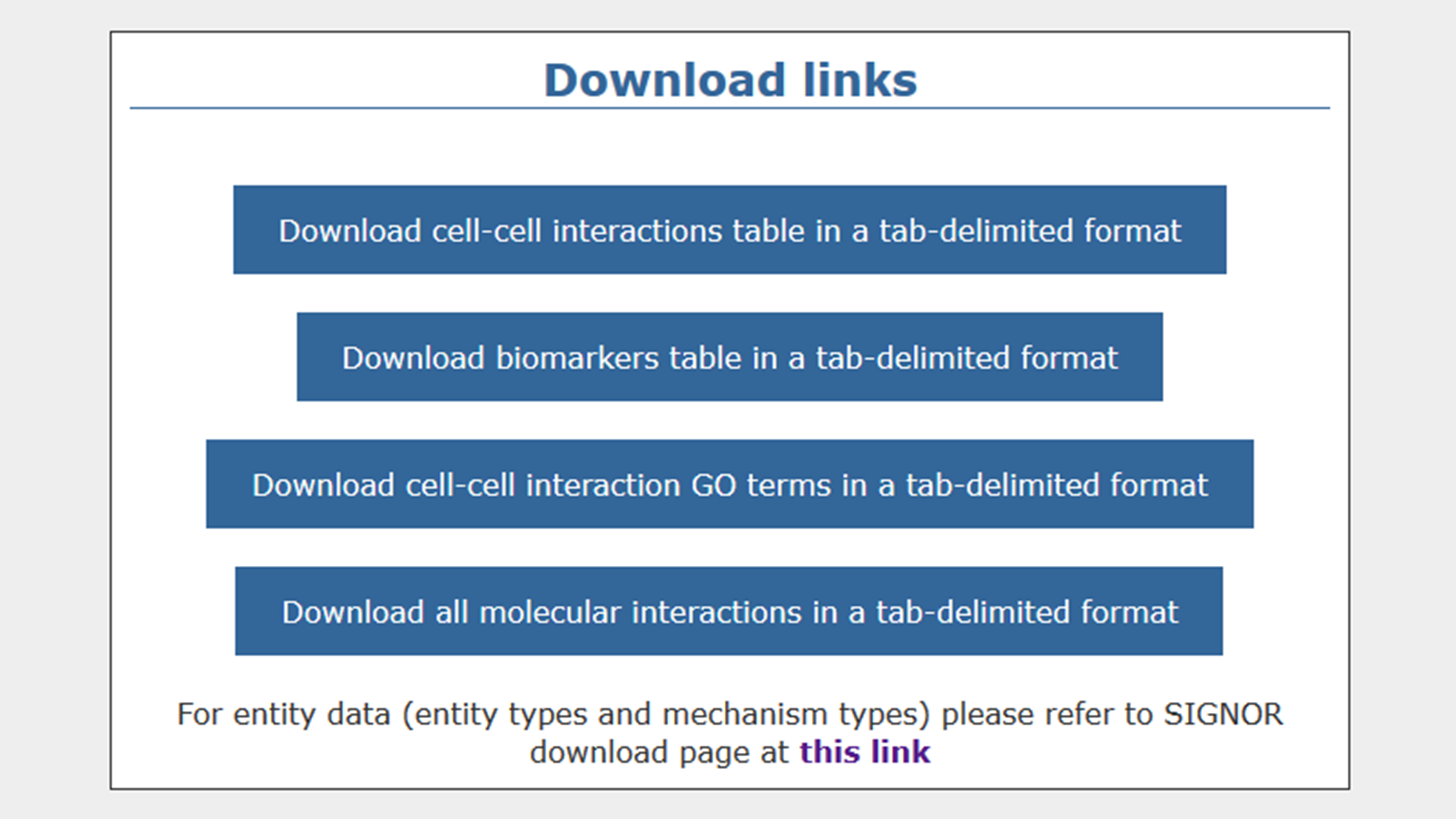
You can download data from the download page. You can choose to download cell-cell interactions, molecular interactions, GO terms for cell-cell interaction effects, and biomarkers. All data can be downloaded in a TAB-delimited format, in order to be opened with softwares of common use.
Searching entities
Users can perform two kinds of search via the entity search form: they can search a molecular entity (1) (gene or protein) by typing the protein/gene name or protein ID (Uniprot ID), or they can search a cellular entity (3) by typing its name (e.g. M1 macrophage, satellite).
A table is shown as result, indicating the matches found on our database. In the molecular entities results table (3), user can choose either to display causal interactions involving the selected entity stored on SIGNOR database, or to display pathways in which that entity is involved.
In the cell result table (4), matches found are shown as well. In this case, by chosing a cell-type, users can display cell-cell interactions in which the selected cell is involved, or pathways for the selected cell type.
Searching articles
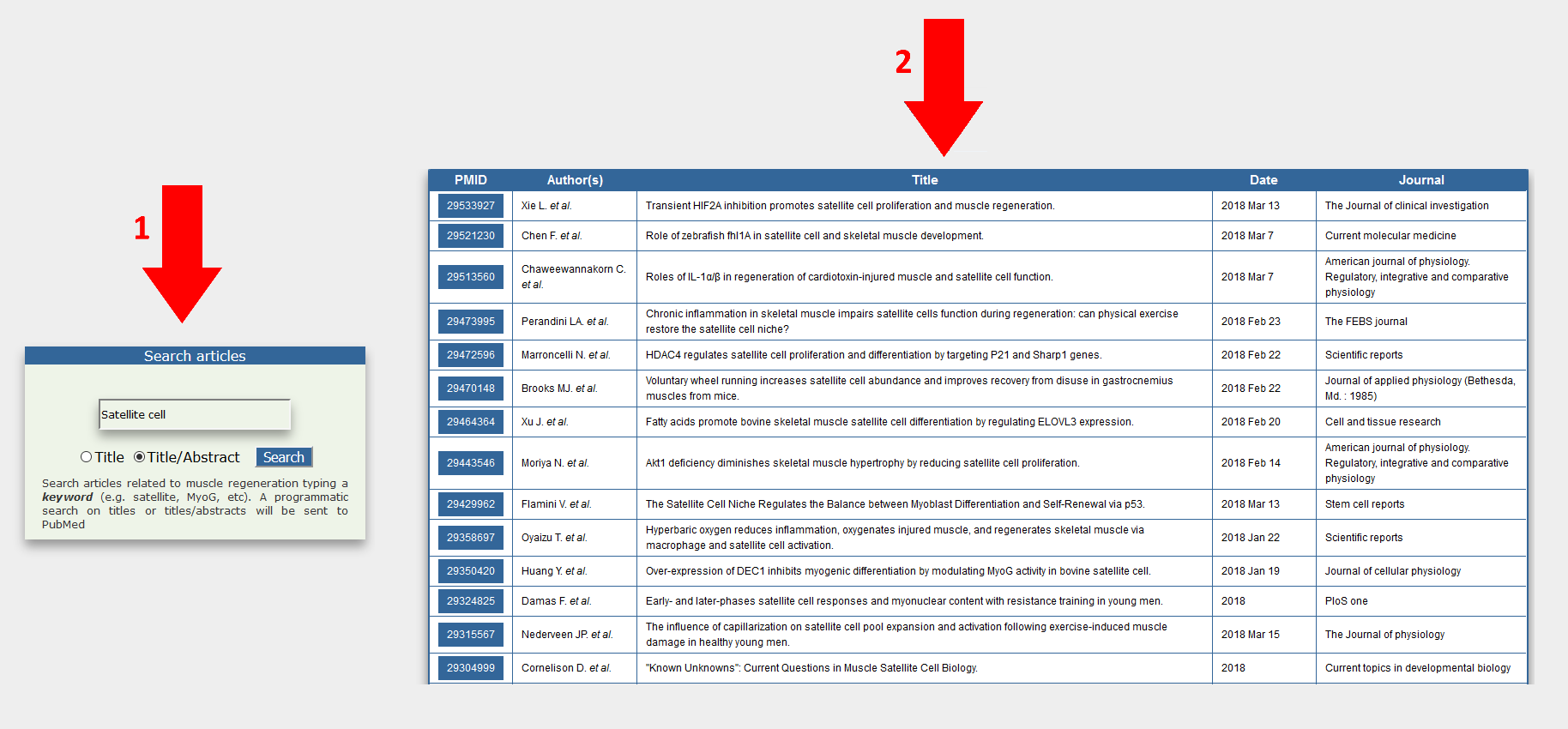
Users are allowed to perform a literature search for PubMed articles related to muscle regeneration by typing keyword(s) on the search article bar (1). A programmatic query is sent to PubMed to retrieve articles whose content is focused on muscle regeneration. Output data is given as a table (2) reporting PubMed IDs (PMIDs) and article titles.
Working with BooleSim models
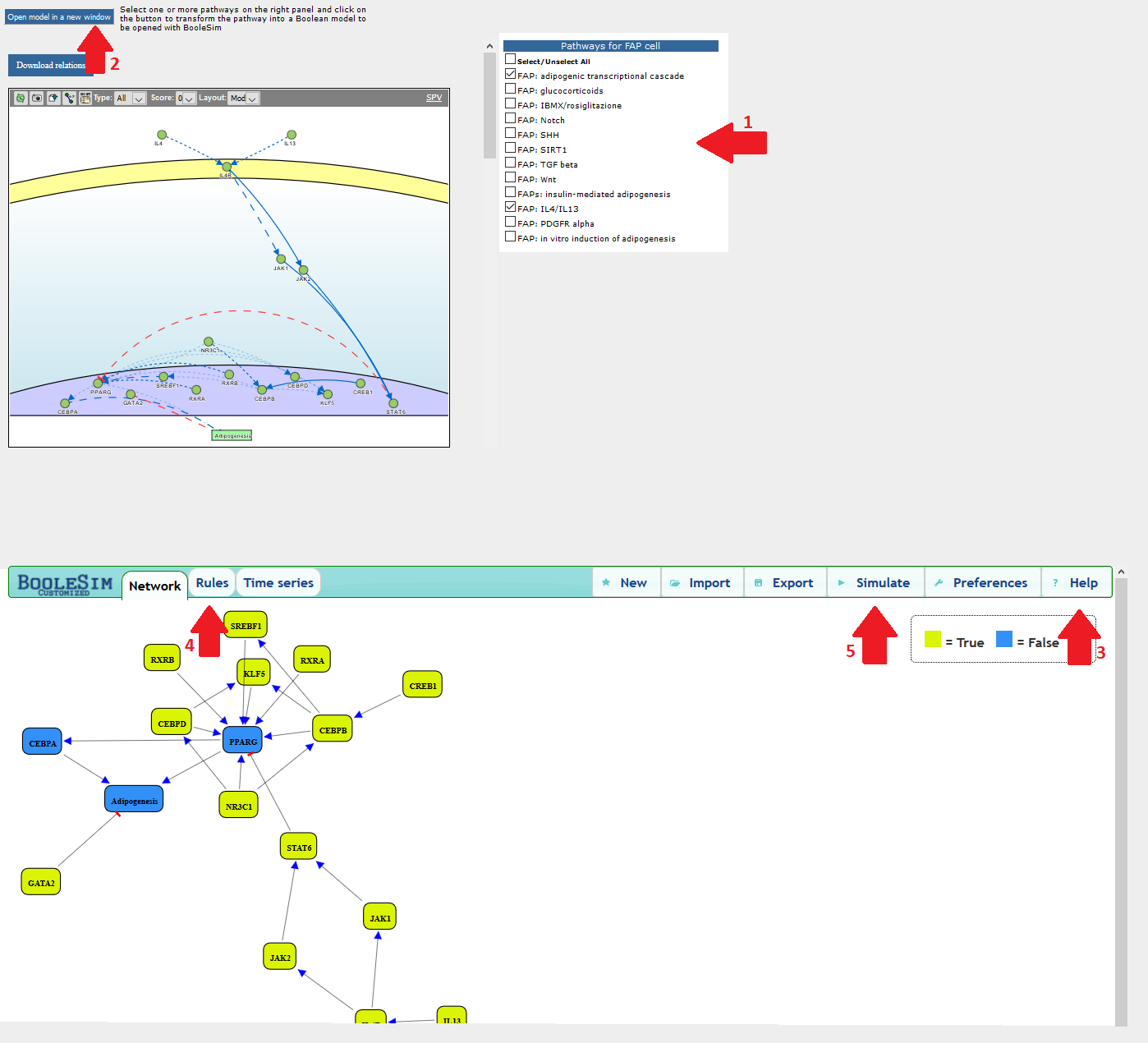
With BooleSim implementation, users can simulate a Boolean network and identify the system equilibrium states. We have implemented a simple algorithm that transforms selected pathways (1) into Boolean networks, which users can directly open in a customized implementation of BooleSim by clicking on the top right button of the pathway panel (2). Once open the BooleSim frame, users can click the “Help” tab for help (3), or navigate to the Boolesim "online help" (https://github.com/matthiasbock/BooleSim/wiki) for more details. You can inspect and, if desirable, change the rules underlying the model by clicking the "Rules" tab (4). By clicking any of the node you can change its state from true to false and vice versa. By clicking the "Simulate" tab (5) the tool changes the state of the nodes according to the underlying rules.